Optimization by Grid search¶
In this section, we will explain how to perform a grid-type search to analyze atomic coordinates from diffraction data.
The grid type search is compatible with MPI.
It is necessary to prepare the data file MeshData.txt that defines the search grid in advance.
Location of the sample files¶
The sample files are located in sample/mapper.
The following files are stored in the folder:
basedirectoryDirectory that contains reference files to proceed with calculations in the main program. The reference files include:
exp.d,rfac.d,tleed4.i, andtleed5.i.input.tomlInput file of the main program.
MeshData.txtData file for the search grid.
ref_ColorMap.txtReference file of the output to check if the calculation is performed correctly.
prepare.sh,do.shScript prepared for bulk calculation of this tutorial.
Below, we will describe these files and then show the actual calculation results.
Reference files¶
tleed4.i, tleed5.i, and rfac.d are the parameter files for satleed.
The atomic coordinates to be optimized should be replaced by the keywords such as opt000 or opt001 in tleed5.i.
exp.d is the reference experimental data.
The search grid is given by MeshData.txt.
In this tutorial, the content of MeshData.txt is as follows:
1 -0.490000 0.777500
2 -0.490000 0.977500
3 -0.490000 1.177500
4 -0.490000 1.377500
5 -0.490000 1.577500
...
The first column denotes the sequential id, and the second and subsequent columns denote the parameter values for opt000 and opt001 in tleed5.i.
Note
A script file make_meshdata.py is prepared to generate MeshData.txt. Run the script as follows:
$ python3 make_meshdata.py > MeshData.txt
Input file¶
This section describes the input file for the main program, input.toml.
The details of input.toml can be found in the input file section of the manual.
The following is the content of input.toml in the sample file.
[base]
dimension = 2
output_dir = "output"
[solver]
name = "leed"
[solver.config]
path_to_first_solver = "../satleed/satl1.exe"
path_to_second_solver = "../satleed/satl2.exe"
[solver.param]
string_list = ["opt000", "opt001"]
[solver.reference]
path_to_base_dir = "./base"
rfactor = "satleed"
[algorithm]
name = "mapper"
label_list = ["z1", "z2"]
[algorithm.param]
mesh_path = "./MeshData.txt"
[runner]
ignore_error = true
First, [base] section is explained.
dimensionis the number of variables to be optimized. In this case, it is2since we are optimizing two variables as described intleed5.i.output_diris the name of directory for the outputs. If it is omitted, the results are written in the directory in which the program is executed.
[solver] section specifies the solver to be used inside the main program and its settings.
nameis the name of the solver you want to use. In this tutorial it isleed.
The solver can be configured in the subsections [solver.config], [solver.param], and [solver.reference].
[solver.config] section specifies options for satl1.exe and satl2.exe that are called from the main program.
path_to_first_solveris the command name ofsatl1.exe. It is specified as a path to the executable file, or searched from the PATH environment variable.path_to_second_solveris the command name ofsatl2.exe. It is specified as a path to the executable file, or searched from the PATH environment variable.
[solver.param] section specifies the parameters for the solver.
string_listspecifies the list of embedded keywords in the input files.
[solver.reference] section specifies the location of the experimental data and the range to read.
path_to_base_dirspecifies the path to the directory for the reference data.rfactorspecifies the definition of the R-factor. The default value isrpe(Pendry’s R-factor). In this example,satleedis specified that uses the output of SATLEED.
[algorithm] section specifies the algorithm to use and its settings.
nameis the name of the algorithm you want to use. In this tutorial we will usemappersince we will be using grid-search method.label_listis a list of label names to be attached to the output ofopt000andopt001.
[algorithm.param] section specifies the parameters for the search algorithm.
mesh_pathis the file name of the search grid.
[runner] section specifies the settings on how to execute the solver.
ignore_errorspecifies how to treat the errors occurred within the execution of the solver. If it is set to true, the result is treated as NaN and the execution of the program is continued.
For details on other parameters that can be specified in the input file, please see the Input File section of the manual.
Calculation execution¶
Before running the sample program, you need to run setup.sh in the sample/satleed directory to compile SATLEED and generate satl1.exe and satl2.exe.
Move to the directory in which sample files are located:
$ cd sample/mapper
Then, run the main program. The computation time will take only a few minutes on a normal PC.
$ mpiexec -np 4 odatse-LEED input.toml | tee log.txt
Here, the calculation using MPI parallel with 4 processes will be done.
When executed, a folder for each MPI rank will be created in output, and the results of the calculations are stored there.
The standard output will be shown like as follows.
name : mapper
label_list : ['z1', 'z2']
param.mesh_path : ./MeshData.txt
Iteration : 1/121
Iteration : 2/121
Iteration : 3/121
Iteration : 4/121
...
z1 and z2 are the candidate parameters for each mesh and R-factor is the function value at that point.
Finally, the R-factor calculated at all the points on the grid will be written to ColorMap.txt.
In this case, the following results will be obtained.
-0.490000 0.777500 0.861000
-0.490000 0.977500 1.004700
-0.490000 1.177500 0.909900
-0.490000 1.377500 0.896600
-0.490000 1.577500 1.009500
-0.490000 1.777500 0.779100
-0.490000 1.977500 0.944200
-0.490000 2.177500 0.966500
-0.490000 2.377500 0.867000
-0.490000 2.577500 0.907000
-0.490000 2.777500 0.924100
-0.390000 0.777500 0.801900
-0.390000 0.977500 0.793900
...
The first and second columns contain the values of opt000 and opt001, and the third column contains the R-factor.
Note that do.sh is available as a script for batch calculation.
In do.sh, the difference between ColorMap.dat and ref_ColorMap.dat is also examined.
Here is what it does, without further explanation.
#!/bin/sh
sh prepare.sh
time mpiexec -np 4 odatse-LEED input.toml
echo diff output/ColorMap.txt ref_ColorMap.txt
res=0
diff output/ColorMap.txt ref_ColorMap.txt || res=$?
if [ $res -eq 0 ]; then
echo TEST PASS
true
else
echo TEST FAILED: ColorMap.txt and ref_ColorMap.txt differ
false
fi
Visualization of calculation results¶
By plotting ColorMap.txt, we can estimate the region where the value of R-factor becomes small.
In this case, the following command will create a two-dimensional parameter space diagram ColorMapFig.png.
$ python3 plot_colormap_2d.py
Looking at the generated figure, we can see that it has the minimum value around (0.0, 1.75).
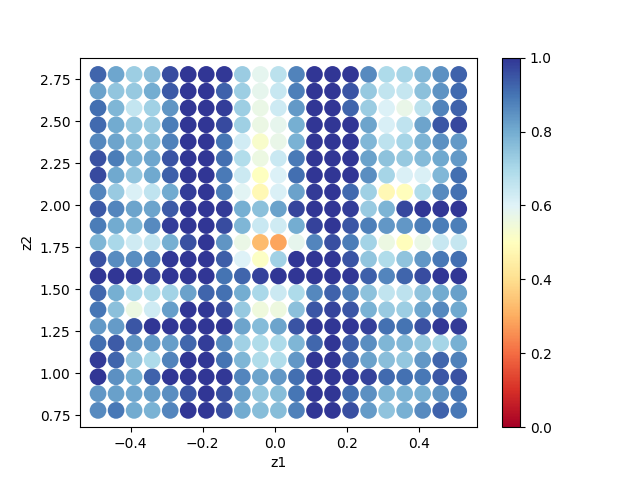
Fig. 1 R-factor on a two-dimensional parameter space (calculated on 21x21 mesh).¶