Optimization by population annealing Monte Carlo method¶
This tutorial describes how to estimate atomic positions from the experimental diffraction data by using the population annealing Monte Carlo method (PAMC).
Sample files¶
Sample files are available from sample/single_beam/pamc .
This directory includes the following files:
bulk.txtThe input file of
bulk.exe.experiment.txt,template.txtReference files for the main program.
ref.txtSolution file for checking whether the calucation is successful or not (reference for
fx.txt).input.tomlThe input file of odatse-STR.
prepare.sh,do.shScript files for running this tutorial.
In the following, we will subscribe these files and then show the result.
Reference files¶
This tutorial uses the reference files, template.txt and experiment.txt,
which are the same as those used in the previous tutorial (Optimization by Nelder-Mead method).
Input files¶
This subsection describes the input file.
For details, see the population annealing Monte Carlo method in ODAT-SE manual.
input.toml in the sample directory is shown as the following:
[base]
dimension = 2
output_dir = "output"
[algorithm]
name = "pamc"
label_list = ["z1", "z2"]
seed = 12345
[algorithm.param]
min_list = [3.0, 3.0]
max_list = [6.0, 6.0]
step_list = [0.3, 0.3]
[algorithm.pamc]
numsteps_annealing = 5
bmin = 0.0
bmax = 200.0
Tnum = 21
Tlogspace = false
nreplica_per_proc = 10
[solver]
name = "sim-trhepd-rheed"
run_scheme = "subprocess"
[solver.config]
cal_number = [1]
[solver.param]
string_list = ["value_01", "value_02" ]
[solver.post]
normalization = "TOTAL"
[solver.reference]
path = "experiment.txt"
exp_number = [1]
In the following, we will briefly describe this input file. For details, see the manual of the population annealing Monte Carlo method in ODAT-SE manual.
[base] section describes the settings for a whole calculation.
dimensionis the number of variables you want to optimize. In this case, specify2because it optimizes two variables.output_diris the name of directory for the outputs. If it is omitted, the results are written in the directory in which the program is executed.
[algorithm] section sets the algorithm to use and its settings.
nameis the name of the algorithm you want to use. In this tutorial we will use PAMC, so specifypamc.label_listis a list of labels to be attached to the output ofvalue_0x(x = 1,2).seedis the seed that a pseudo-random number generator uses.
[algorithm.param] section sets the parameter space to be explored.
min_listis a lower bound andmax_listis an upper bound.unit_listis step length in one MC update (deviation of Gaussian)
[algorithm.pamc] section sets the parameters for PAMC.
numsteps_annealingis the number of interval steps between temperature decreasing.bmin,bmaxare the minimum and the maximum of inversed temperature, respectively.Tnumis the number of (inversed) temperature points.When
Tlogspaceistrue, the temperature points are distributed uniformly in the logarithmic space.nreplica_per_procis the number of replicas (MC walkers) in one MPI process.
[solver] section specifies the solver to use inside the main program and its settings.
See the Optimization by Nelder-Mead method tutorial.
Calculation¶
First, move to the folder where the sample file is located. (Hereinafter, it is assumed that you are the root directory of odatse-STR.)
$ cd sample/single_beam/pamc
Copy bulk.exe and surf.exe as in the tutorial for the direct problem.
$ cp ../../sim-trhepd-rheed/src/bulk.exe .
$ cp ../../sim-trhepd-rheed/src/surf.exe .
Execute bulk.exe to generate bulkP.b.
$ ./bulk.exe
Then, run the main program. It will take a few secondes on a normal PC.
$ mpiexec -np 4 odatse-STR input.toml | tee log.txt
Here, the calculation is performed using MPI parallel with 4 processes.
If you are using OpenMPI and you request more processes than the number of available CPU cores, add the --oversubscribed option to the mpiexec command.
When executed, a folder for each MPI rank will be created under the directory output.
There, trial_TXXX.txt and result_TXXX.txt will be created (where XXX is the index of points).
The former contains the parameters evaluated in each Monte Carlo step and the value of the objective function at each temperature, and the latter contains the parameters actually adopted.
These files are concatnated into trial.txt and result.txt, respectively.
These files have the same format: the first two columns are time (step) and the index of walker in the process, the third is the (inversed) temperature, the fourth column is the value of the objective function, and the fifth and subsequent columns are the parameters. The final two columns are the weight of walker (Neal-Jarzynski weight) and the index of the grand ancestor (the replica index at the beginning of the calculation).
# step walker beta fx z1 z2 weight ancestor
0 0 0.0 0.07702743614780189 5.788848278451443 3.949126663745358 1.0 0
0 1 0.0 0.08737730661436376 3.551756435031283 3.6136808356591192 1.0 1
0 2 0.0 0.04954470587051104 4.70317508724506 4.786634108937754 1.0 2
0 3 0.0 0.04671675601156148 5.893543559206865 4.959531290614713 1.0 3
0 4 0.0 0.04142014655238446 5.246719912601735 4.960709612555206 1.0 4
In the case of the sim-trhepd-rheed solver, a subfolder LogXXXX_YYYY (XXXX is the index of MC steps) is created under each working directory, and the rocking curve information and other outputs are recorded.
best_result.txt is filled with the information about the parameters with the value of the optimal objective function (R-factor), the rank from which it was obtained, and the Monte Carlo step.
nprocs = 4
rank = 0
step = 71
walker = 5
fx = 0.008186713312593607
z1 = 4.225633749839847
z2 = 5.142666117413409
Finally, fx.txt stores the statistics at each temperature point:
# $1: 1/T
# $2: mean of f(x)
# $3: standard error of f(x)
# $4: number of replicas
# $5: log(Z/Z0)
# $6: acceptance ratio
0.0 0.06428002079611472 0.002703413400677839 40 0.0 0.795
10.0 0.061399304916174735 0.002649424392996749 40 -0.6280819199879947 0.85
20.0 0.05904248889111052 0.0031622711212952034 40 -1.2283060742855603 0.74
30.0 0.04956921148431115 0.0028298565759159633 40 -1.7991035905899855 0.67
...
The first column is (inversed) temperature, and the second/third ones are the mean and standard error of \(f(x)\), respectively. The fourth column is the number of replicas and the fifth one is the logarithm of the ratio of the partition functions, \(\log(Z_n/Z_0)\), where \(Z_0\) is the partition function at the first temperature. The sixth column is the acceptance ratio of MC updates.
In addition, do.sh is prepared as a script for batch calculation.
do.sh also checks the difference between best_result.txt and ref.txt.
The content of the script is shown below, though further information will be omitted.
#!/bin/sh
sh prepare.sh
./bulk.exe
time mpiexec --oversubscribe -np 4 odatse-STR input.toml
echo diff output/fx.txt ref.txt
res=0
diff output/fx.txt ref.txt || res=$?
if [ $res -eq 0 ]; then
echo TEST PASS
true
else
echo TEST FAILED: output/fx.txt and ref.txt differ
false
fi
Visualization¶
By illustrating result_T.txt, you can estimate regions where the parameters with small R-factor are.
In this case, the figure result_fx.png and result_T.png of the 2D parameter space is created by using the following command.
The color of symbols of result_fx.png and result_T.png mean R-factor and \(\beta\), respectively.
$ python3 plot_result_2d.py
Looking at the resulting diagram, we can see that the samples are concentrated near (5.25, 4.25) and (4.25, 5.25), and that the R-factor value is small there.
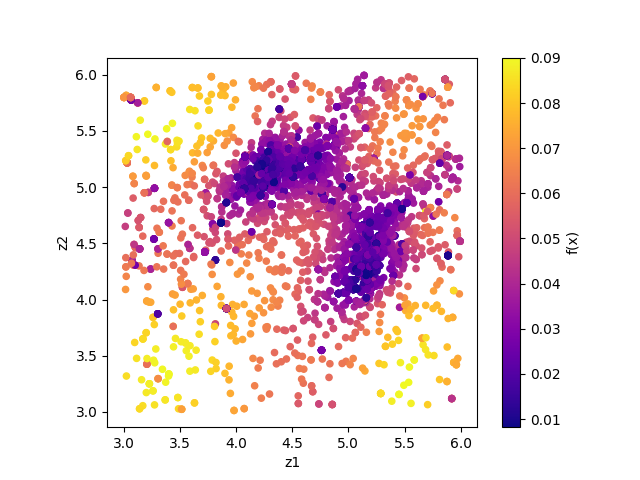
Fig. 6 Sampled parameters and R-factor. The horizontal axes is x1 (value_01) and the vertical axes is x2 (value_02).¶
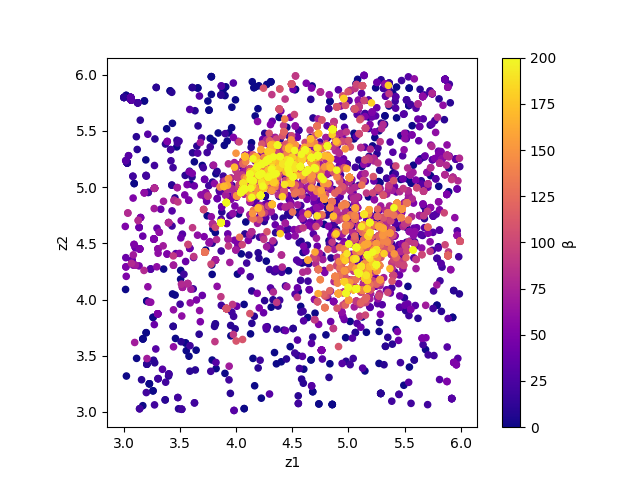
Fig. 7 Sampled parameters and \(\beta\). The horizontal axes is x1 (value_01) and the vertical axes is x2 (value_02).¶
When generate_rocking_curve in [solver] section is set to true, RockingCurve.txt is stored in each subfolder, LogXXXX_YYYY (XXXX is an index of MC step and YYYY is an index of a replica in the MPI process).
By using this, it is possible to compare with the experimental value according to the procedure of the previous tutorial.